Calculating a Geometry Matrix using the Voxel Framework¶
In this demonstration, we calculate the geometry matrix for a slightly more complicated bolometer system, consisting of 3 cameras each of which have 16 foils looking through a single aperture. For simplicity we discretise the measurement space into toroidally symmetric voxels of identical rectangular cross section, although it is possible to have an arbitrary cross section in the voxel framework.
In addition to the geometry matrix, a regularisation operator is generated. This is needed for regularised tomographic inversions, as shown in the Inversion with Voxels demo. We use a simple Laplacian operator as the regularisation operator, which corresponds to isotropic smoothing.
"""
This example demonstrates calculating the geometry matrix for a
bolometer system using a voxel collection. In this instance the voxel
collection make up of a regular grid of rectangular voxels, but this is
not strictly necessary: it just makes deriving a regularisation operator
for the inversions in other demos a bit easier.
"""
import math
from pathlib import Path
import pickle
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
import numpy as np
from raysect.core import Node, Point2D, Point3D, Vector3D, rotate_basis, rotate_y, translate
from raysect.core.math.function.float import Arg2D
from raysect.optical import World
from raysect.optical.material import AbsorbingSurface
from raysect.primitive import Box, Cylinder, Subtract
from cherab.tools.inversions import ToroidalVoxelGrid
from cherab.tools.observers import BolometerCamera, BolometerSlit, BolometerFoil
# Convenient constants
XAXIS = Vector3D(1, 0, 0)
YAXIS = Vector3D(0, 1, 0)
ZAXIS = Vector3D(0, 0, 1)
ORIGIN = Point3D(0, 0, 0)
# Bolometer geometry
BOX_WIDTH = 0.05
BOX_WIDTH = 0.2
BOX_HEIGHT = 0.07
BOX_DEPTH = 0.2
SLIT_WIDTH = 0.004
SLIT_HEIGHT = 0.005
FOIL_WIDTH = 0.0013
FOIL_HEIGHT = 0.0038
FOIL_CORNER_CURVATURE = 0.0005
SLIT_SENSOR_SEPARATION = 0.1
FOIL_SEPARATION = 0.00508 # 0.2 inch between foils
SENSOR_ANGLES = [-18, -6, 6, 18]
world = World()
########################################################################
# Make a simple vessel geometry
########################################################################
centre_column_radius = 1
vessel_wall_radius = 3.7
vessel_height = 3.7
vessel = Subtract(
Cylinder(radius=vessel_wall_radius, height=vessel_height),
Cylinder(radius=centre_column_radius, height=vessel_height),
material=AbsorbingSurface(), name="Vessel", parent=world,
)
########################################################################
# Build a simple bolometer system
########################################################################
def make_bolometer_camera():
# The camera consists of a box with a rectangular slit and 4 sensors,
# each of which has 4 foils.
# In its local coordinate system, the camera's slit is located at the
# origin and the sensors below the z=0 plane, looking up towards the slit.
camera_box = Box(lower=Point3D(-BOX_WIDTH / 2, -BOX_HEIGHT / 2, -BOX_DEPTH),
upper=Point3D(BOX_WIDTH / 2, BOX_HEIGHT / 2, 0))
# Hollow out the box
inside_box = Box(lower=camera_box.lower + Vector3D(1e-5, 1e-5, 1e-5),
upper=camera_box.upper - Vector3D(1e-5, 1e-5, 1e-5))
camera_box = Subtract(camera_box, inside_box)
# The slit is a hole in the box
aperture = Box(lower=Point3D(-SLIT_WIDTH / 2, -SLIT_HEIGHT / 2, -1e-4),
upper=Point3D(SLIT_WIDTH / 2, SLIT_HEIGHT / 2, 1e-4))
camera_box = Subtract(camera_box, aperture)
camera_box.material = AbsorbingSurface()
# Instance of the bolometer camera
bolometer_camera = BolometerCamera(camera_geometry=camera_box)
# The bolometer slit in this instance just contains targeting information
# for the ray tracing, since we have already given our camera a geometry
# The slit is defined in the local coordinate system of the camera
slit = BolometerSlit(slit_id="Example slit", centre_point=ORIGIN,
basis_x=XAXIS, dx=SLIT_WIDTH, basis_y=YAXIS, dy=SLIT_HEIGHT,
parent=bolometer_camera)
for j, angle in enumerate(SENSOR_ANGLES):
# 4 bolometer foils, spaced at equal intervals along the local X axis
# The bolometer positions and orientations are given in the local coordinate
# system of the camera, just like the slit
sensor = Node(name="Bolometer sensor", parent=bolometer_camera,
transform=rotate_y(angle) * translate(0, 0, -SLIT_SENSOR_SEPARATION))
# The foils are shifted relative to the centre of the sensor by -1.5, -0.5, 0.5 and 1.5
# times the foil-foil separation
for i, shift in enumerate([-1.5, -0.5, 0.5, 1.5]):
# Note that the foils will be parented to the camera rather than the
# sensor, so we need to define their transform relative to the camera
foil_transform = sensor.transform * translate(shift * FOIL_SEPARATION, 0, 0)
foil = BolometerFoil(detector_id="Foil {} sensor {}".format(i + 1, j + 1),
centre_point=ORIGIN.transform(foil_transform),
basis_x=XAXIS.transform(foil_transform), dx=FOIL_WIDTH,
basis_y=YAXIS.transform(foil_transform), dy=FOIL_HEIGHT,
slit=slit, parent=bolometer_camera, units="Power",
accumulate=False, curvature_radius=FOIL_CORNER_CURVATURE)
bolometer_camera.add_foil_detector(foil)
return bolometer_camera
# Make several cameras distributed around the outside of the vessel
camera_angles = [20, 60, 100]
rotation_origin = Point2D(1.5, 1.5)
cameras = []
for camera_angle in camera_angles:
camera = make_bolometer_camera()
camera.transform = (translate(rotation_origin.x, 0, rotation_origin.y)
* rotate_y(camera_angle)
* translate(0, 0, 2.0)
* rotate_basis(-ZAXIS, YAXIS)
)
camera.parent = world
camera.name = "Angle {}".format(camera_angle)
cameras.append(camera)
########################################################################
# Show the lines of sight of all the bolometer channels
########################################################################
def _point3d_to_rz(point):
return Point2D(math.hypot(point.x, point.y), point.z)
fig, ax = plt.subplots()
all_foils = [foil for camera in cameras for foil in camera]
for foil in all_foils:
slit_centre = foil.slit.centre_point
slit_centre_rz = _point3d_to_rz(slit_centre)
ax.plot(slit_centre_rz[0], slit_centre_rz[1], 'ko')
origin, hit, _ = foil.trace_sightline()
centre_rz = _point3d_to_rz(foil.centre_point)
ax.plot(centre_rz[0], centre_rz[1], 'kx')
origin_rz = _point3d_to_rz(origin)
hit_rz = _point3d_to_rz(hit)
ax.plot([origin_rz[0], hit_rz[0]], [origin_rz[1], hit_rz[1]], 'k')
ax.add_patch(Rectangle((centre_column_radius, 0), edgecolor='k', facecolor='none',
width=(vessel_wall_radius - centre_column_radius),
height=vessel_height,))
ax.axis('equal')
ax.set_title("Bolometer camera lines of sight")
ax.set_xlabel("r")
ax.set_ylabel("z")
########################################################################
# Define the region of interest for the inversions
########################################################################
# The inversions will be performed on the emission profile used in the
# radiation_function.py demo, so we'll trim the voxel grid down to the
# emitting region
PLASMA_AXIS = Point2D(1.5, 1.5)
LCFS_RADIUS = 1
# distance of the virtual inner wall from LCFS
WALL_LCFS_OFFSET = 0.1
# Build a mask, only including cells within the wall
# We'll use raysect's function framework for this
radius_squared = ((Arg2D('x') - PLASMA_AXIS.x)**2 + (Arg2D('y') - PLASMA_AXIS.y)**2)
mask = radius_squared <= (WALL_LCFS_OFFSET + LCFS_RADIUS)**2
########################################################################
# Produce a voxel grid
########################################################################
print("Producing the voxel grid...")
# We'll use a grid of rectangular voxels here, all of which are the same
# size. Neither the shape nor the uniform size are required for using the
# voxels, but it makes this example a bit simpler.
# Define the centres of each voxel, as an (nx, ny, 2) array
nx = 40
ny = 60
cell_centres = np.meshgrid(np.linspace(1, 3, nx), np.linspace(0, 3, ny))
cell_r = np.linspace(1, 3, nx)
cell_z = np.linspace(0, 3, ny)
cell_r_grid, cell_z_grid = np.broadcast_arrays(cell_r[:, None], cell_z[None, :])
cell_centres = np.stack((cell_r_grid, cell_z_grid), axis=-1) # (nx, ny, 2) array
cell_dx = cell_centres[1, 0] - cell_centres[0, 0]
cell_dy = cell_centres[0, 1] - cell_centres[0, 0]
# Define the positions of the vertices of the voxels, as an (nx, ny, 4, 2) array
cell_vertex_displacements = np.asarray([-0.5 * cell_dx - 0.5 * cell_dy,
-0.5 * cell_dx + 0.5 * cell_dy,
0.5 * cell_dx + 0.5 * cell_dy,
0.5 * cell_dx - 0.5 * cell_dy])
all_cell_vertices = np.swapaxes(cell_centres[..., None], -2, -1) + cell_vertex_displacements
# Produce a (ncells, nvertices, 2) array of coordinates to initialise the
# ToroidalVoxelCollection. Here, ncells = number of cells inside mask,
# nvertices = 4. The ToroidalVoxelGrid expects a flat list of (nvertices, 2)
# arrays to define voxels, since there is no implicit assumption that the voxels
# lie on a grid.
enclosed_cells = []
grid_mask = np.empty((nx, ny), dtype=bool)
grid_index_2D_to_1D_map = {}
grid_index_1D_to_2D_map = {}
# Identify the cells that are enclosed by the polygon,
# simultaneously write out grid mask and grid map.
unwrapped_cell_index = 0
for ix in range(nx):
for iy in range(ny):
# p1, p2, p3, p4 = cell_vertices[ix][iy]
vertices = all_cell_vertices[ix, iy]
# if any points are inside the polygon, retain this cell
if any(mask(p[0], p[1]) for p in vertices):
grid_mask[ix, iy] = True
# We'll need these maps for generating the regularisation operator
grid_index_2D_to_1D_map[(ix, iy)] = unwrapped_cell_index
grid_index_1D_to_2D_map[unwrapped_cell_index] = (ix, iy)
enclosed_cells.append(vertices)
unwrapped_cell_index += 1
else:
grid_mask[ix, iy] = False
num_cells = len(enclosed_cells)
voxel_data = np.empty((num_cells, 4, 2)) # (number of cells, 4 coordinates, x and y values)
for i, row in enumerate(enclosed_cells):
p1, p2, p3, p4 = row
voxel_data[i, 0, :] = p1
voxel_data[i, 1, :] = p2
voxel_data[i, 2, :] = p3
voxel_data[i, 3, :] = p4
voxel_grid = ToroidalVoxelGrid(voxel_data)
########################################################################
# Produce a regularisation operator for inversions
########################################################################
# We'll use simple isotropic smoothing here, in which case an ND second
# derivative operator (the laplacian operator) is appropriate
grid_laplacian = np.zeros((num_cells, num_cells))
for ith_cell in range(num_cells):
# get the 2D mesh coordinates of this cell
ix, iy = grid_index_1D_to_2D_map[ith_cell]
neighbours = 0
try:
n1 = grid_index_2D_to_1D_map[ix - 1, iy] # neighbour 1
except KeyError:
pass
else:
grid_laplacian[ith_cell, n1] = -1
neighbours += 1
try:
n2 = grid_index_2D_to_1D_map[ix - 1, iy + 1] # neighbour 2
except KeyError:
pass
else:
grid_laplacian[ith_cell, n2] = -1
neighbours += 1
try:
n3 = grid_index_2D_to_1D_map[ix, iy + 1] # neighbour 3
except KeyError:
pass
else:
grid_laplacian[ith_cell, n3] = -1
neighbours += 1
try:
n4 = grid_index_2D_to_1D_map[ix + 1, iy + 1] # neighbour 4
except KeyError:
pass
else:
grid_laplacian[ith_cell, n4] = -1
neighbours += 1
try:
n5 = grid_index_2D_to_1D_map[ix + 1, iy] # neighbour 5
except KeyError:
pass
else:
grid_laplacian[ith_cell, n5] = -1
neighbours += 1
try:
n6 = grid_index_2D_to_1D_map[ix + 1, iy - 1] # neighbour 6
except KeyError:
pass
else:
grid_laplacian[ith_cell, n6] = -1
neighbours += 1
try:
n7 = grid_index_2D_to_1D_map[ix, iy - 1] # neighbour 7
except KeyError:
pass
else:
grid_laplacian[ith_cell, n7] = -1
neighbours += 1
try:
n8 = grid_index_2D_to_1D_map[ix - 1, iy - 1] # neighbour 8
except KeyError:
pass
else:
grid_laplacian[ith_cell, n8] = -1
neighbours += 1
grid_laplacian[ith_cell, ith_cell] = neighbours
########################################################################
# Calculate the geometry matrix for the grid
########################################################################
print("Calculating the geometry matrix...")
# The voxel grid must be in the same world as the bolometers
voxel_grid.parent = world
sensitivity_matrix = []
for camera in cameras:
for foil in camera:
print("Calculating sensitivity for {}...".format(foil.name))
sensitivity_matrix.append(foil.calculate_sensitivity(voxel_grid))
sensitivity_matrix = np.asarray(sensitivity_matrix)
# Plot the sensitivity matrix, summed over all foils
fig, ax = plt.subplots()
voxel_grid.plot(ax=ax, title="Total sensitivity [m³sr]",
voxel_values=sensitivity_matrix.sum(axis=0))
ax.set_xlabel("r")
ax.set_ylabel("z")
# Save the voxel grid information and the geometry matrix for use in other demos
voxel_grid_data = {'voxel_data': voxel_data, 'laplacian': grid_laplacian,
'grid_index_1D_to_2D_map': grid_index_1D_to_2D_map,
'grid_index_2D_to_1D_map': grid_index_2D_to_1D_map,
'sensitivity_matrix': sensitivity_matrix}
script_dir = Path(__file__).parent
with open(script_dir / "voxel_grid_data.pickle", "wb") as f:
pickle.dump(voxel_grid_data, f)
plt.show()
Caption The lines of sight of the 48 foils in 3 bolometer cameras, used to generate the sensitivity matrix.¶
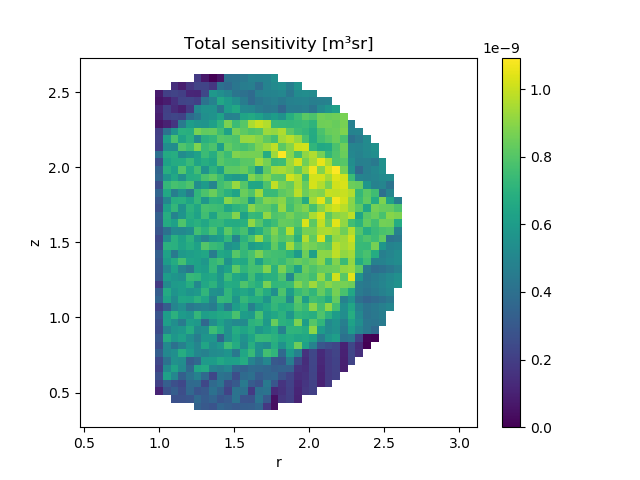
Caption The result of summing the sensitivty of all foils, for each voxel in the voxel collection.¶