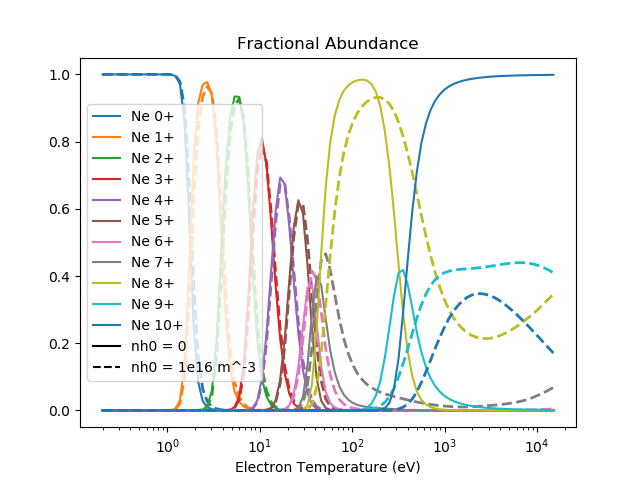
Fractional Abundances¶
Some example code for requesting fractional abundances and plotting them.
import numpy as np
import matplotlib.pyplot as plt
from cherab.core.atomic import neon, hydrogen
from cherab.openadas import OpenADAS
from scipy.optimize import lsq_linear
def get_rates_recombination(element):
"""
load recombinatio rates for all ionic stages
"""
coef_recom = {}
for i in np.arange(1, elem.atomic_number + 1):
coef_recom[i] = adas.recombination_rate(element, int(i))
return coef_recom
def get_rates_tcx(donor, donor_charge, receiver):
"""
load thermal charge-exchange recombination rates for all ionic stages
"""
coef_tcx = {}
for i in np.arange(1, elem.atomic_number + 1):
coef_tcx[i] = adas.thermal_cx_rate(donor, donor_charge, receiver, int(i))
return coef_tcx
def get_rates_ionisation(element):
"""
load ionisation rates for all ionic stages
:param element:
:return:
"""
coef_ionis = {}
for i in np.arange(0, elem.atomic_number):
coef_ionis[i] = adas.ionisation_rate(element, int(i))
return coef_ionis
def solve_ion_balance(element, n_e, t_e, coef_ion, coef_recom, nh0=None, coef_tcx=None):
atomic_number = element.atomic_number
# construct the fractional abundance matrix
matbal = np.zeros((atomic_number + 1, atomic_number + 1))
matbal[0, 0] -= coef_ion[0](n_e, t_e)
matbal[0, 1] += coef_recom[1](n_e, t_e)
matbal[-1, -1] -= coef_recom[atomic_number](n_e, t_e)
matbal[-1, -2] += coef_ion[atomic_number - 1](n_e, t_e)
if nh0 is not None:
matbal[0, 1] += nh0 / n_e * coef_tcx[1](n_e, t_e)
matbal[-1, -1] -= nh0 / n_e * coef_tcx[atomic_number](n_e, t_e)
for i in range(1, atomic_number):
matbal[i, i - 1] += coef_ion[i - 1](n_e, t_e)
matbal[i, i] -= (coef_ion[i](n_e, t_e) + coef_recom[i](n_e, t_e))
matbal[i, i + 1] += coef_recom[i + 1](n_e, t_e)
if nh0 is not None:
matbal[i, i] -= nh0 / n_e * coef_tcx[i](n_e, t_e)
matbal[i, i + 1] += nh0 / n_e * coef_tcx[i + 1](n_e, t_e)
# for some reason calculation of stage abundance seems to yield better results than calculation of fractional abun.
matbal = matbal * ne # multiply by ne to calculate abundance instead of fractional abundance
# add sum constraints. Sum of all stages should be equal to electron density
matbal = np.concatenate((matbal, np.ones((1, matbal.shape[1]))), axis=0)
# construct RHS of the balance steady-state equation
rhs = np.zeros((matbal.shape[0]))
rhs[-1] = ne
abundance = lsq_linear(matbal, rhs, bounds=(0, ne))["x"]
# normalize to ne to get fractional abundance
frac_abundance = abundance / ne
return frac_abundance
# initialise the atomic data provider
adas = OpenADAS(permit_extrapolation=True)
elem = neon
temperature_steps = 100
ne = 1E19
nh0 = 1e15
numstates = elem.atomic_number + 1
# Collect rate coefficients
rates_ion = get_rates_ionisation(elem)
rates_recom = get_rates_recombination(elem)
rates_tcx = get_rates_tcx(hydrogen, 0, elem)
electron_temperatures = [10 ** x for x in np.linspace(np.log10(rates_recom[1].raw_data["te"].min()),
np.log10(rates_recom[1].raw_data["te"].max()),
num=temperature_steps)]
# calculate ionization balance for electron temperature rates
ion_balance = np.zeros((elem.atomic_number + 1, len(electron_temperatures)))
ion_balance_tcx = np.zeros((elem.atomic_number + 1, len(electron_temperatures)))
for j, te in enumerate(electron_temperatures):
ion_balance[:, j] = solve_ion_balance(elem, ne, te, rates_ion, rates_recom)
ion_balance_tcx[:, j] = solve_ion_balance(elem, ne, te, rates_ion, rates_recom, nh0, rates_tcx)
# plot results
for i in range(elem.atomic_number + 1):
try:
ionisation_rates = [rates_ion[i](1E19, x) for x in electron_temperatures]
plt.loglog(electron_temperatures, ionisation_rates, '-x', label='{0} {1}+'.format(elem.symbol, i))
except KeyError:
continue
plt.ylim(1E-21, 1E-10)
plt.legend()
plt.xlabel("Electron Temperature (eV)")
plt.title("Ionisation Rates")
plt.figure()
for i in range(elem.atomic_number + 1):
try:
recombination_rates = [rates_recom[i](1E19, x) for x in electron_temperatures]
plt.loglog(electron_temperatures, recombination_rates, '-x', label='{0} {1}+'.format(elem.symbol, i))
except KeyError:
continue
plt.ylim(1E-21, 1E-10)
plt.legend()
plt.xlabel("Electron Temperature (eV)")
plt.title("Recombination Rates")
plt.figure()
for i in range(elem.atomic_number + 1):
try:
tcx_rates = [rates_tcx[i](1E19, x) for x in electron_temperatures]
plt.loglog(electron_temperatures, tcx_rates, '-x', label='{0} {1}+'.format(elem.symbol, i))
except KeyError:
continue
plt.ylim(1E-21, 1E-10)
plt.legend()
plt.xlabel("Electron Temperature (eV)")
plt.title(" Thermal Charge-Exchange Recombination Rates")
plt.figure()
for i in range(elem.atomic_number + 1):
pl = plt.semilogx(electron_temperatures, ion_balance[i, :], label='{0} {1}+'.format(elem.symbol, i))
plt.semilogx(electron_temperatures, ion_balance_tcx[i, :], '--',
color=pl[0].get_color(), lw=2)
plt.plot([], [], "k-", label="nh0 = 0")
plt.plot([], [], "k--", label="nh0 = 1e16 m^-3")
plt.xlabel("Electron Temperature (eV)")
plt.title('Fractional Abundance')
plt.legend()
plt.show()